What is metaproteomics?
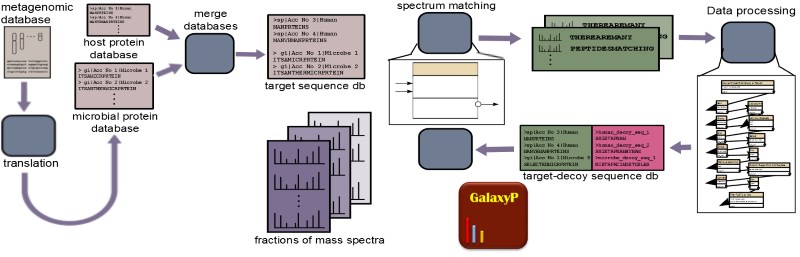
Metaproteomics is the use of mass spectrometry (MS)-based proteomics to study the collective proteins expressed by microbial communities, also known as microbiomes. These communities may be part of environmental samples (e.g. soil, waste water etc) or contained within a host organism (e.g. human gut, oral cavity etc). Metaproteomics complements metagenomics, which classifies the type (taxonomy) and abundance of different microbial species present in samples of interest. The profile of expressed proteins offered by metaproteomics provides a more direct measure of the functional state of the community. It has been suggested that the functional profile offered by metaproteomics may give a more accurate depiction of the dynamics and response of microbial communities as compared to profiling the taxonomic changes.
Galaxy-P is well-suited for metaproteomics data analysis, as this approach requires the use of many different software programs, including those used in metagenomics and MS-based proteomics, integrated into workflows. Galaxy-P has created an educational instance with training materials for metaproteomics research. You can access z.umn.edu/metaproteomicsgateway to access tools and workflows related to metaproteomics research.
The Galaxy-P team has published several seminal papers demonstrating the value of the platform for metaproteomics.
- Protein relative abundance patterns associated with sucrose-induced dysbiosis are conserved across taxonomically diverse oral microcosm biofilm models of dental caries. Rudney JD, Jagtap PD, Reilly CS, Chen R, Markowski TW, Higgins L, Johnson JE, Griffin TJ. (2015) 3:69. PubMed link
- Metaproteomic analysis using the Galaxy framework. Jagtap PD, Blakely A, Murray K, Stewart S, Kooren J, Johnson JE, Rhodus NL, Rudney J, Griffin TJ. (2015) 15:3553-65. PubMed link
- Multi-omic data analysis using Galaxy. Boekel J, Chilton JM, Cooke IR, Horvatovich PL, Jagtap PD, Käll L, Lehtiö J, Lukasse P, Moerland PD, Griffin TJ. Nat Biotechnol. (2015) 33:137-9. PubMed link
- Members of the Galaxy-P team also co-edited a special issue on Microbiomes and metaproteomics in the journal Proteomics, which can be found here.
