What is proteogenomics?
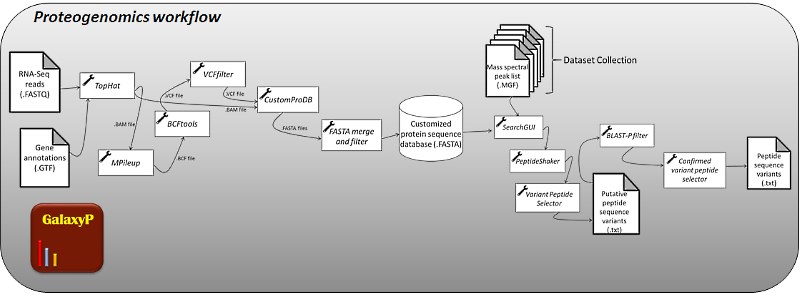
Proteogenomics integrates genomic information with mass spectrometry (MS)-based proteomics data. The most common workflow uses transcriptome sequencing information, obtained using next-generation sequencing methods like RNA-seq. The assembled transcriptome is translated in-silico to generate a database of possible proteins expressed in the sample. This database includes those proteins with possible novel sequences, derived from DNA or RNA sequence variants. Matching proteomics data (in the form of tandem mass spectra, also know as MS/MS spectra) with sequences in the database provides a confirmation of the expression of novel protein sequences in the sample. The proteogenomics approach provides new insights into novel protein sequences that may carry new functions and be drivers of disease. This approach also provides a powerful means to annotate genomes.
Galaxy-P provides an ideal platform for proteogenomics, which requires integration of software for analysis of genomic or transcriptomic data (e.g. RNA-seq data) and also MS-based proteomics data. Galaxy-P has created an educational instance with training materials for proteogenomics research. You can access z.umn.edu/proteogenomicsgateway to access tools and workflows related to protegenomics research.
The Galaxy P-team has published several seminal papers on the use of Galaxy for proteogenomics.
- Proteogenomic Analysis of a Hibernating Mammal Indicates Contribution of Skeletal Muscle Physiology to the Hibernation Phenotype. Anderson KJ, Vermillion KL, Jagtap P, Johnson JE, Griffin TJ, Andrews MT. J Proteome Res. (2016) 15:1253-61. PubMed link
- Characterizing Cardiac Molecular Mechanisms of Mammalian Hibernation via Quantitative Proteogenomics. Vermillion KL, Jagtap P, Johnson JE, Griffin TJ, Andrews MT. J Proteome Res. (2015) 14:4792-804. PubMed link (see also this commentary in Chemical and Engineering News)
- Flexible and accessible workflows for improved proteogenomic analysis using the Galaxy framework. Jagtap PD, Johnson JE, Onsongo G, Sadler FW, Murray K, Wang Y, Shenykman GM, Bandhakavi S, Smith LM, Griffin TJ. J Proteome Res. (2014) 13:5898-908. PubMed link
- Using Galaxy-P to leverage RNA-Seq for the discovery of novel protein variations. Sheynkman GM, Johnson JE, Jagtap PD, Shortreed MR, Onsongo G, Frey BL, Griffin TJ, Smith LM. BMC Genomics. (2014) 15:703. PubMed link
